
Sanger's DNA Squencing
ABOUT HIM...
-Born in Rendcombe, England
- conscientious objector during the war
- Study in Cambridge University
- Discover stucture of protein
- Novel Prize winner in year 1958
-Inventor of Sanger DNA squencing

After his Ph.D. in 1943, Sanger started working for A. C. Chibnall, on identifying the free amino groups in insulin. In the course of identifying the amino groups, Sanger figured out ways to order the amino acids. He was the first person to obtain a protein sequence. By doing so, Sanger proved that proteins were ordered molecules and by analogy, the genes and DNA that make these proteins should have an order or sequence as well. Sanger won his first Nobel Prize for Chemistry in 1958 for his work on the structure of protein.
By 1951, Sanger was on the staff of the Medical Research Council at Cambridge University. In 1962, he moved with the Medical Research Council to the Laboratory of Molecular Biology in Cambridge where Francis Crick, John Kendrew, Aaron Klug and others were all working on a DNA-related problem. Solving the problem of DNA sequencing became a natural extension of his work in protein sequencing. Sanger initially investigated ways to sequence RNA because it was smaller. Eventually, this led to techniques that were applicable to DNA and finally to the dideoxy method most commonly used in sequencing reactions today. Sanger won a second Nobel Prize for Chemistry in 1980 sharing it with Walter Gilbert, for their contributions concerning the determination of base sequences in nucleic acids, and Paul Berg for his work on recombinant DNA.
During the 1970s, Frederick Sanger developed a new technique allowing the base sequence of DNA to be determined. The design of his method is still very popular today. Sanger employed dideoxynucleic acids, ddNTPs, in addition to deoxynucleic acids together in the amplification of DNA during the Polymerase Chain Reaction (PCR). Instead of amplifying a section of DNA, the ddNTPS would cause amplification to terminate in a random number of amplified products. The ddNTPs were also radioactively labeled for detection. The end result of the Sanger chemistry would be a series of new DNA products with sizes increasing by a single base.
The PCR products were separated on a medium, polyacrylamide gel, with smaller products migrating faster than the larger products. The end result appeared like a ladder when detected.The basics of the chemistry remain the same today. What has changed in Sanger sequencing is the method of separating the products and detection.
In 1992, the Wellcome Trust and the Medical Research Council established the Sanger Centre, a research center for furthering the knowledge of genomes. The Sanger Centre is one of the main sequencing centers of the Human Genome Sequencing Project and sequencing projects of other organisms are also underway at the Sanger Centre.
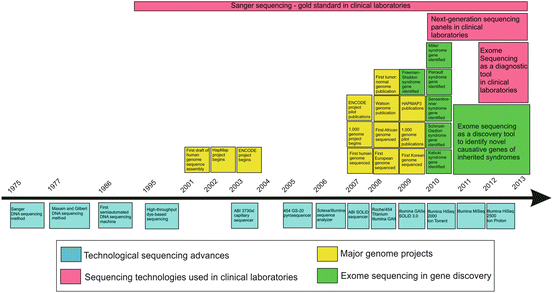
Timeline of the major advances in sequencing technologies starting with Sanger sequencing and ending with the next-generation–sequencing (blue).